An Adaptive Pipeline From Scientific Data to Models
Under DARPA’s Synergistic Discovery and Design program, a team composed of scientists from Duke, Rutgers, Montana State, and Florida Atlantic Universities, as well as Geometric Data Analytics, and Netrias, Inc., broadly researched and developed data driven techniques for scientific discovery and robust design, proving feasibility through program challenge problems with Yeast States, Novel Chassis, Protein Stability, and Perovskite.
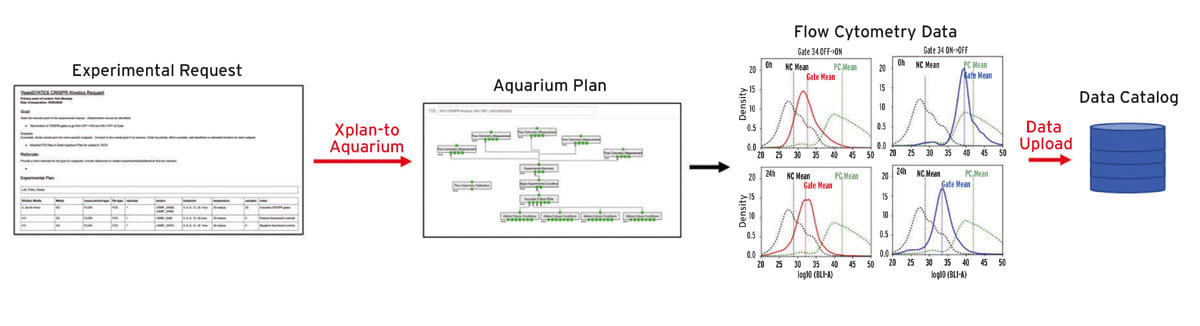
The Duke Team, composed of scientists from Duke University, Rutgers The State University of New Jersey (Rutgers), Montana State University, Florida Atlantic University, Geometric Data Analytics (GDA), and Netrias, Inc., has worked broadly within the Defense Advanced Research Projects Agency (DARPA) Synergistic Discovery and Design (SD2) program, contributing to efforts in the Yeast States, Novel Chassis, Protein Stability, and Perovskite challenge problems (CP).
The SD2 program was structured across five technical areas (TAs), TA1 - Data-Centric Scientific Discovery, TA2 – Design in the Context of Uncertainty, TA3 – Hypothesis and Design Evaluation, TA4 – Data and Analysis Hub, and TA5 – Challenge Problem Integrator, where the Duke Team supported TA1 and TA3 capabilities. Early in the program it was realized that some of the data needed to achieve goals of this effort could not be produced by the automated and semi-automated TA3 laboratories. To merge the data collected in the benchtop laboratory of the Duke Team with the automated labs, it was necessary to utilize approaches for protocol execution and data collection that were utilized by the TA3 labs. In collaboration with the University of Washington (UW) Biofab team, Aquarium for the benchtop was developed, enabling the Duke Team’s benchtop lab to execute protocols and collect data, that when delivered to the database, was indistinguishable from data collected at the automated TA3 labs.
Before SD2, circuit designs in synthetic biology were focused on proposed functions. Findings early in the program indicated that synthetic genetic circuits did not perform functions consistently across varying growth conditions that likely alter parameters in which the circuit functions. At that time there were no tools for designing circuits taking into account both function and robustness to growth conditions. The practical importance of robust designs lies in the ability of circuits to perform their functions in the face of varying conditions of a deployment, even within highly controlled conditions such as a fermenter.
Dynamic Signatures Generated by Regulatory Networks (DSGRN) was developed as a design tool that could computationally assess the robustness of a particular circuit topology across parameter conditions. As well, with circuit performance data, DSGRN could infer the mode of failure of circuits so that they might be “repaired”.
DSGRN has been wrapped with extensive tooling to improve the design process, especially for logic circuits with arbitrary biological parts. Multiple design problems were tackled in Yeast States and Novel Chassis including the comprehensive analysis of all three node networks for bistability, the redesign of 2-input logic circuits for enhanced robustness, the redesign of a 3- input logic circuit for glitch removal, and the analysis of experimental data from an external Department of Defense (DoD) partner. The design process was reduced from months to days with 50-100 percent qualitative matching to data where it was available.
Several SD2 achievements facilitated this work. An easy-to-use design tool was created that predicts the robustness of logic circuit designs given user-supplied experimental constraints and network functionality requirements. The concepts of robustness of performance incorporating design parameters, neighboring parameters, and continuation in Hill function models were improved. Concrete connections between DSGRN parameters were developed as well as build constraints that are easily communicated to experimentalists. The number of DSGRN-computable network topologies was increased, including more complex networks (more in-edges, multiple edges between nodes, self-repressors, no in-edges, no out-edges, non-monotone interactions).
This work was performed by Steven B. Haase, Ph.D., Duke University, for the Air Force Research Laboratory. For more information, download the Technical Support Package (free white paper) below.
This Brief includes a Technical Support Package (TSP).
An Adaptive Pipeline From Scientific Data to Models
(reference AFRL-2023059) is currently available for download from the TSP library.
Don't have an account?
Overview
The document titled "An Adaptive Pipeline from Scientific Data to Models" is a final technical report produced by a collaborative team from Duke University and several other institutions, sponsored by the Air Force Research Laboratory and DARPA. Covering the period from September 2017 to September 2022, the report outlines the development of data-driven techniques aimed at enhancing scientific discovery and robust design, particularly within the context of synthetic biology.
The report emphasizes the integration of various tools and methodologies to create a nearly fully automated Design, Build, Test, Learn (DBTL) loop, which is crucial for accelerating research and development processes. It highlights the importance of compatibility among different tools, which significantly contributed to the success of the program, particularly in the Yeast States challenge problem.
Key sections of the report include discussions on data acquisition, which merges high-throughput automated experimentation with low-throughput benchtop data acquisition, and the robustness of synthetic biology systems. The document also details tools developed for automated data pre-processing, normalization, and quality control, as well as tools for data aggregation and acceleration of the DBTL loop. A notable tool mentioned is the Build Request Parser, which automates the workflow by parsing build information from semi-structured documents.
Metrics for success are outlined, focusing on improvements in speed, accuracy, and volume of data processing. The report also discusses the transition of these tools and methodologies to address urgent needs, such as accelerating COVID-19 therapeutic interventions and vaccine development.
In conclusion, the report reflects on the lessons learned during the COVID-19 pandemic, emphasizing the importance of having a robust infrastructure for national bio-preparedness. The findings and methodologies presented in this report are intended to facilitate future research and development efforts in synthetic biology and related fields, ultimately contributing to advancements in scientific knowledge and practical applications. The document serves as a comprehensive resource for understanding the integration of data-driven approaches in scientific research and the potential for these methodologies to drive innovation.
Top Stories
INSIDERAerospace
![]() How Airbus is Using w-DED to 3D Print Larger Titanium Airplane Parts
How Airbus is Using w-DED to 3D Print Larger Titanium Airplane Parts
NewsAutomotive
![]() Microvision Aquires Luminar, Plans Relationship Restoration, Multi-industry Push
Microvision Aquires Luminar, Plans Relationship Restoration, Multi-industry Push
INSIDERManned Systems
![]() A Next Generation Helmet System for Navy Pilots
A Next Generation Helmet System for Navy Pilots
INSIDERDesign
![]() New Raytheon and Lockheed Martin Agreements Expand Missile Defense Production
New Raytheon and Lockheed Martin Agreements Expand Missile Defense Production
ArticlesManned Systems
![]() Accelerating Down the Road to Autonomy
Accelerating Down the Road to Autonomy
NewsPower
![]() Ford Announces 48-Volt Architecture for Future Electric Truck
Ford Announces 48-Volt Architecture for Future Electric Truck
Webcasts
Electronics & Computers
![]() Cooling a New Generation of Aerospace and Defense Embedded...
Cooling a New Generation of Aerospace and Defense Embedded...
Test & Measurement
![]() Battery Abuse Testing: Pushing to Failure
Battery Abuse Testing: Pushing to Failure
Internet of Things
![]() A FREE Two-Day Event Dedicated to Connected Mobility
A FREE Two-Day Event Dedicated to Connected Mobility
Unmanned Systems
![]() Quiet, Please: NVH Improvement Opportunities in the Early Design Cycle
Quiet, Please: NVH Improvement Opportunities in the Early Design Cycle
Transportation
![]() Advantages of Smart Power Distribution Unit Design for Automotive &...
Advantages of Smart Power Distribution Unit Design for Automotive &...
Aerospace
![]() Sesame Solar's Nanogrid Tech Promises Major Gains in Drone Endurance
Sesame Solar's Nanogrid Tech Promises Major Gains in Drone Endurance